Biography
Lorena is a plant pathologist in the epidemiology group within the Cell and Molecular Sciences department. Her research focuses on the foliar microbiome of agricultural crops or their wild progenitors and how phytopathogenesis impacts community structure. In particular, she is interested in investigating the potential to exploit the foliar and endophytic microbiome for tailored disease control of plant pathogenic bacteria and fungi.
Research
Current Research Interests and Impact
Native microbial strains are better adapted to offering plants protection from local biotic stresses, and profiling these communities offers an approach for biocontrol research in any crop pathosystem. Using a crop agnostic approach, cataloguing the native plant microbiome has the potential to provide low-risk biological control alternatives and a foundation for commercial application.
Foliar Microbiome Research
-
- Phyllosphere (epiphyte & endophtye) characterisation of healthy/diseased signatures
- Assessing function of hub taxa for crop protection
- Biocontrol of plant pathogenic bacteria and fungi
- Work with industry towards creation of safe biologicals

Secondary Metabolite Research
- Discovery of SM involvement in phytopathogenesis
- Identification of cross-kingdom SM communication
- Validating function of SM gene clusters
- Improve crop protection through plant resistance inducers
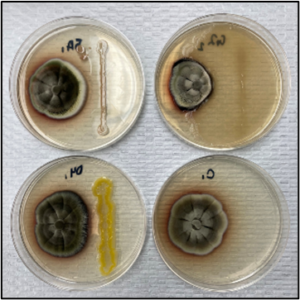
Past research
Previous Experience
2019-2023 Post-doctoral Research Plant Pathologist, USDA-ARS, Fargo, North Dakota, USA
2019 Ph.D. Plant Pathology – University of California, Davis, California, USA
2012 M.S. Plant Pathology – Oregon State University, Corvallis, Oregon, USA
2010 B.S. Microbiology minor Chemistry – Texas State University, San Marcos, Texas, USA
Publications
Fredell, G., Bienkowski, D., Light, S.E. and Rangel, L.I., 2025. Distribution, potential commercial adoption, and possible impacts of the endophytic fungus, Epichloë, in UK pasture grasses and cereals. CABI Plant Health Cases. https://doi.org/10.1079/planthealthcases.2025.0002
Rangel, L.I., Wyatt, N., Courneya, I., Natwick, M.B., Secor, G.A., Rivera-Varas, V., & M.D. Bolton. 2024. CbCyp51 mediated DMI resistance is modulated by codon bias. Phytopathology. https://doi.org/10.1094/PHYTO-01-24-0034-R
Rangel, L.I. & J.H.L. Leveau. 2024. Applied microbiology of the phyllosphere. Applied Microbiology and Biotechnology. 108: 211. https://doi.org/10.1007/s00253-024-13042-4
Rangel, L.I. & M.D. Bolton. 2022. The unsung roles of microbial secondary metabolite effectors in the plant disease cacophony. Current Opinion in Plant Biology. 68: 102233. https://doi.org/10.1016/j.pbi.2022.102233
Rangel, L.I., Hamilton, O., de Jonge, R., & M.D Bolton. 2021. Fungal social influencers: secondary metabolites as a platform for shaping the plant-associated community. The Plant Journal. 3: 632-645. https://doi.org/10.1111/tpj.15490
Ebert, M.K.*, Rangel, L.I.*, Spanner, R.E., Taliadoros, D., Wang, X., Friesen, T.L., de Jonge, R., Neubauer, J.D., Secor, G.A., Thomma, B.P.H.J., Stukenbrock, E.H. & M.D. Bolton. 2020. Identification and characterization of Cercospora beticola necrosis-inducing effector CbNIP1. Molecular Plant Pathology. 00:1–16. https://doi.org/10.1111/mpp.13026
Rangel, L.I., Spanner, R., Ebert, M.K., Pethybridge, S., Stukenbrock, E., de Jonge, R., Secor, G.A. & M.D. Bolton. 2020. Cercospora beticola: the intoxicating lifestyle of the leaf spot pathogen of sugar beet. Molecular Plant Pathology. 21:1020-1041. https://doi.org/10.1111/mpp.12962
Rangel, L.I., Henkels, M.D., Shaffer, B.T., Walker, F.L., Davis II, E.W., Stockwell, V.O., Bruck, D., Taylor, B.J. and J.E. Loper. 2016. Characterization of toxin complex gene clusters and insect toxicity of bacteria representing four subgroups of Pseudomonas fluorescens. PLoS One. 11(8), p.e0161120. https://doi.org/10.1371/journal.pone.0161120
Loper, J.E., Henkels, M.D., Rangel, L.I., Olcott, M.H., Walker, F.L., Bond, K.L., Kidarsa, T.A., Hesse, C.N., Sneh, B., Stockwell, V.O. and B.J. Taylor. 2016. Rhizoxin analogs, orfamide A and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environmental Microbiology. 18(10): 3509-3521. https://doi.org/10.1111/1462-2920.13369
Henkels, M.D., Kidarsa, T.A., Shaffer, B.T, Goebel, N.C., Burlinson, P., Mavrodi, D.V., Bentley, M.A., Rangel, L.I., Davis, E.W., Thomashow, L.S., Zabriskie, T.M., Preston, G.M. and J.E. Loper. 2014. Pseudomonas protegens Pf-5 causes discoloration and pitting of mushroom caps due to the production of antifungal metabolites. Molecular Plant-Microbe Interactions. 27: 733-746. https://doi.org/10.1094/MPMI-10-13-0311-R
Loper, J.E., Hassan, K.A., Mavrodi, D.V., Davis, E.W., Lim, C.K., Shaffer, B.T., Elbourne, L.D.H., Hartney, S., Stockwell, V., Breakwell, K., Henkels, M.D., Tetu, S.G., Rangel, L.I., Wilson, N.L., vanMortel, J., Song, C., Blumhagen, R., Radune, D., Hostetler, J., Brinkac, L., Durkin, S., Kluepfel, D., Wechter, P., Anderson, A., Kim, Y.C., Pierson, L.S., Pierson, E.A., Lindow, S.E., Raaijmakers, J.M., Weller, D., Thomashow, L., Allen, A. and I.T. Paulsen. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genetics. 8: e1002784. https://doi.org/10.1371/journal.pgen.1002784
